| Entry |
|
|---|
| Name |
Cloxacillin benzathine (USP) |
|---|
| Formula |
(C19H18ClN3O5S)2. C16H20N2
|
|---|
| Exact mass |
1110.2938
|
|---|
| Mol weight |
1112.106
|
|---|
| Structure |
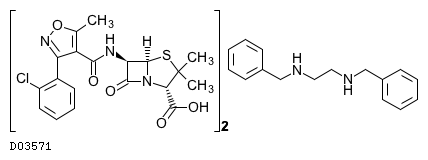
|
|---|
| Class |
Antibacterial
DG01710 beta-Lactam antibiotic
DG01713 Penicillin skeleton group
DG01480 Penicillin
DG01779 beta-Lactamase resistant penicillin
|
|---|
| Remark |
|
|---|
| Efficacy |
Antibacterial, Cell wall biosynthesis inhibitor |
|---|
| Target |
penicillin binding protein |
|---|
| Pathway |
|
|---|
| Interaction |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
J ANTIINFECTIVES FOR SYSTEMIC USE
J01 ANTIBACTERIALS FOR SYSTEMIC USE
J01C BETA-LACTAM ANTIBACTERIALS, PENICILLINS
J01CF Beta-lactamase resistant penicillins
J01CF02 Cloxacillin
D03571 Cloxacillin benzathine (USP)
Drug groups [BR:br08330]
Antibacterial
DG01710 beta-Lactam antibiotic
DG01713 Penicillin skeleton group
DG01480 Penicillin
DG00541 Cloxacillin
D03571 Cloxacillin benzathine
DG01779 beta-Lactamase resistant penicillin
DG00541 Cloxacillin
D03571 Cloxacillin benzathine
Antimicrobials [BR:br08307]
Antibacterials
Cell wall biosynthesis inhibitor, beta-lactam
beta-Lactamase resistant penicillin
D03571 Cloxacillin benzathine (USP)
Drug groups [BR:br08330]
Antibacterial
DG01710 beta-Lactam antibiotic
DG01713 Penicillin skeleton group
DG01480 Penicillin
DG00541 Cloxacillin
DG01779 beta-Lactamase resistant penicillin
DG00541 Cloxacillin
Antimicrobials abbreviations [BR:br08327]
Antibacterials
Cell wall biosynthesis inhibitor, beta-lactam
beta-Lactamase resistant penicillin
DG00541 Cloxacillin
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 76
1 C1y C 25.1447 -10.9212
2 C5x C 25.1447 -12.3126
3 N1y N 26.5362 -12.3126
4 C1y C 26.5362 -10.9212
5 C1y C 27.8581 -12.7300
6 C1z C 28.6931 -11.6168
7 S2x S 27.8581 -10.5037
8 C1a C 29.6670 -12.5909
9 C1a C 29.6670 -10.6428
10 C6a C 28.3452 -14.1215
11 O6a O 29.7365 -14.1215
12 O6a O 27.5103 -15.2347
13 N1b N 23.9621 -10.2254
14 C5a C 22.7793 -10.9212
15 O5x O 23.9621 -13.0084
16 O5a O 22.7793 -12.3126
17 C8y C 21.5269 -10.2254
18 C8y C 21.5269 -8.8341
19 O2x O 20.2035 -8.4040
20 N5x N 19.3857 -9.5296
21 C8y C 20.2035 -10.6555
22 C1a C 22.6527 -8.0161
23 C8y C 20.2035 -12.0834
24 C8y C 18.9507 -12.8071
25 C8x C 18.9509 -14.2349
26 C8x C 20.1875 -14.9487
27 C8x C 21.4404 -14.2251
28 C8x C 21.4402 -12.7972
29 X Cl 17.7188 -12.0960
30 C8x C 32.1692 -10.1006
31 C8x C 32.1692 -11.5028
32 C8x C 33.3835 -12.2039
33 C8y C 34.5978 -11.5028
34 C8x C 34.5978 -10.1006
35 C8x C 33.3835 -9.3996
36 C1b C 35.8308 -12.2149
37 N1b N 37.0379 -11.5181
38 C1b C 38.2220 -12.2020
39 C1b C 39.4178 -11.5116
40 N1b N 40.6076 -12.1986
41 C1b C 41.8005 -11.5099
42 C8y C 42.9917 -12.1977
43 C8x C 42.9917 -13.6058
44 C8x C 44.2061 -14.3069
45 C8x C 45.4203 -13.6058
46 C8x C 45.4203 -12.1977
47 C8x C 44.2061 -11.4967
48 C1y C 25.1447 -10.9212
49 C5x C 25.1447 -12.3126
50 N1y N 26.5362 -12.3126
51 C1y C 26.5362 -10.9212
52 S2x S 27.8581 -10.5037
53 C1z C 28.6931 -11.6168
54 C1y C 27.8581 -12.7300
55 C6a C 28.3452 -14.1215
56 O6a O 29.7365 -14.1215
57 O6a O 27.5103 -15.2347
58 C1a C 29.6670 -12.5909
59 C1a C 29.6670 -10.6428
60 O5x O 23.9621 -13.0084
61 N1b N 23.9621 -10.2254
62 C5a C 22.7793 -10.9212
63 O5a O 22.7793 -12.3126
64 C8y C 21.5269 -10.2254
65 C8y C 21.5269 -8.8341
66 O2x O 20.2035 -8.4040
67 N5x N 19.3857 -9.5296
68 C8y C 20.2035 -10.6555
69 C8y C 20.2035 -12.0834
70 C8y C 18.9507 -12.8071
71 C8x C 18.9509 -14.2349
72 C8x C 20.1875 -14.9487
73 C8x C 21.4404 -14.2251
74 C8x C 21.4402 -12.7972
75 X Cl 17.7188 -12.0960
76 C1a C 22.6527 -8.0161
BOND 83
1 30 31 2
2 31 32 1
3 32 33 2
4 33 34 1
5 34 35 2
6 30 35 1
7 33 36 1
8 36 37 1
9 37 38 1
10 38 39 1
11 39 40 1
12 40 41 1
13 41 42 1
14 42 43 2
15 43 44 1
16 44 45 2
17 45 46 1
18 46 47 2
19 42 47 1
20 1 2 1
21 2 3 1
22 3 4 1
23 1 4 1
24 3 5 1
25 5 6 1
26 6 7 1
27 4 7 1
28 6 8 1
29 6 9 1
30 5 10 1 #Down
31 10 11 1
32 10 12 2
33 1 13 1 #Up
34 13 14 1
35 2 15 2
36 14 16 2
37 14 17 1
38 17 18 2
39 18 19 1
40 19 20 1
41 20 21 2
42 17 21 1
43 18 22 1
44 21 23 1
45 23 24 2
46 24 25 1
47 25 26 2
48 26 27 1
49 27 28 2
50 23 28 1
51 24 29 1
52 48 49 1
53 49 50 1
54 50 51 1
55 48 51 1
56 50 54 1
57 54 53 1
58 53 52 1
59 51 52 1
60 53 58 1
61 53 59 1
62 54 55 1 #Down
63 55 56 1
64 55 57 2
65 48 61 1 #Up
66 61 62 1
67 49 60 2
68 62 63 2
69 62 64 1
70 64 65 2
71 65 66 1
72 66 67 1
73 67 68 2
74 64 68 1
75 65 76 1
76 68 69 1
77 69 70 2
78 70 71 1
79 71 72 2
80 72 73 1
81 73 74 2
82 69 74 1
83 70 75 1
BRACKET 1 17.2900 -15.8200 17.2900 -7.3500
1 31.2200 -7.3500 31.2200 -15.8200
1 2
ORIGINAL 1 1 2 3 4 7 6 5 10 11 12 8 9 13 16 14 15
1 17 18 19 20 21 22 24 25 26 27 28 29 30 23
REPEAT 1 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64
1 65 66 67 68 69 70 71 72 73 74 75 76 77 78
|
|---|