| Entry |
|
|---|
| Name |
Nebivolol (USAN/INN) |
|---|
| Formula |
C22H25F2NO4
|
|---|
| Exact mass |
405.1752
|
|---|
| Mol weight |
405.43
|
|---|
| Structure |
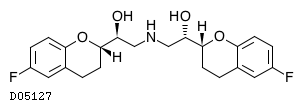
|
|---|
| Simcomp |
|
|---|
| Class |
Cardiovascular agent
DG01466 Adrenergic receptor antagonist
DG01464 beta-Adrenergic receptor antagonist (Beta blocker)
DG01461 beta1-Adrenergic receptor antagonist
DG03231 Antihypertensive
Metabolizing enzyme substrate
DG01644 CYP2D6 substrate
|
|---|
| Remark |
| Product (DG00319): | D06622<US> |
|
|---|
| Efficacy |
Antihypertensive, Vasodilator, beta1-Adrenergic receptor antagonist |
|---|
| Target |
|
|---|
| Pathway |
| hsa04080 | Neuroactive ligand-receptor interaction |
| hsa04261 | Adrenergic signaling in cardiomyocytes |
|
|---|
| Metabolism |
Enzyme: CYP2D6 [HSA: 1565]
|
|---|
| Interaction |
|
|---|
| Brite |
Anatomical Therapeutic Chemical (ATC) classification [BR:br08303]
C CARDIOVASCULAR SYSTEM
C07 BETA BLOCKING AGENTS
C07A BETA BLOCKING AGENTS
C07AB Beta blocking agents, selective
C07AB12 Nebivolol
D05127 Nebivolol (USAN/INN)
Drug groups [BR:br08330]
Cardiovascular agent
DG01466 Adrenergic receptor antagonist
DG01464 beta-Adrenergic receptor antagonist (Beta blocker)
DG01461 beta1-Adrenergic receptor antagonist
DG00319 Nebivolol
D05127 Nebivolol
DG03231 Antihypertensive
DG00319 Nebivolol
D05127 Nebivolol
Metabolizing enzyme substrate
DG01644 CYP2D6 substrate
DG00319 Nebivolol
D05127 Nebivolol
Drug classes [BR:br08332]
Cardiovascular agent
DG03231 Antihypertensive
D05127 Nebivolol
Target-based classification of drugs [BR:br08310]
G Protein-coupled receptors
Rhodopsin family
Adrenaline
ADRB1
D05127 Nebivolol (USAN/INN)
Drug metabolizing enzymes and transporters [br08309.html]
Drug metabolizing enzymes
D05127
Drug groups [BR:br08330]
Cardiovascular agent
DG01466 Adrenergic receptor antagonist
DG01464 beta-Adrenergic receptor antagonist (Beta blocker)
DG01461 beta1-Adrenergic receptor antagonist
DG00319 Nebivolol
DG03231 Antihypertensive
DG00319 Nebivolol
Metabolizing enzyme substrate
DG01644 CYP2D6 substrate
DG00319 Nebivolol
|
|---|
| Other DBs |
|
|---|
| LinkDB |
|
|---|
| KCF data |
ATOM 29
1 C8x C 10.1744 -16.2075
2 C8y C 10.1744 -17.6104
3 C8x C 11.3893 -18.3119
4 C8y C 12.6044 -17.6104
5 C8y C 12.6044 -16.2075
6 C8x C 11.3893 -15.5060
7 C1x C 13.8193 -18.3119
8 C1x C 15.0343 -17.6104
9 C1y C 15.0343 -16.2075
10 O2x O 13.8193 -15.5060
11 C1c C 16.2344 -15.5145
12 X F 8.9594 -18.3119
13 C1b C 17.4252 -16.2019
14 O1a O 16.2345 -14.1033
15 N1b N 18.6185 -15.5129
16 C1b C 19.8106 -16.2011
17 C1c C 21.0032 -15.5125
18 C1y C 22.1958 -16.2009
19 O1a O 21.0033 -14.1032
20 C1x C 22.1959 -17.6102
21 C1x C 23.4109 -18.3115
22 C8y C 24.6258 -17.6099
23 C8y C 24.6256 -16.2007
24 O2x O 23.4106 -15.4994
25 C8x C 25.8408 -18.3113
26 C8y C 27.0557 -17.6097
27 C8x C 27.0556 -16.2005
28 C8x C 25.8406 -15.4992
29 X F 28.2890 -18.3218
BOND 32
1 1 2 2
2 2 3 1
3 3 4 2
4 4 5 1
5 5 6 2
6 1 6 1
7 4 7 1
8 7 8 1
9 8 9 1
10 9 10 1
11 5 10 1
12 9 11 1
13 2 12 1
14 11 13 1
15 11 14 1 #Up
16 13 15 1
17 15 16 1
18 16 17 1
19 17 18 1
20 17 19 1 #Down
21 18 20 1
22 20 21 1
23 21 22 1
24 22 23 1
25 23 24 1
26 18 24 1
27 22 25 2
28 25 26 1
29 26 27 2
30 27 28 1
31 23 28 2
32 26 29 1
|
|---|