| Entry |
|
|---|
| Name |
Antigen processing and presentation - Homo sapiens (human) |
|---|
| Class |
Organismal Systems; Immune system
|
|---|
| Pathway map |
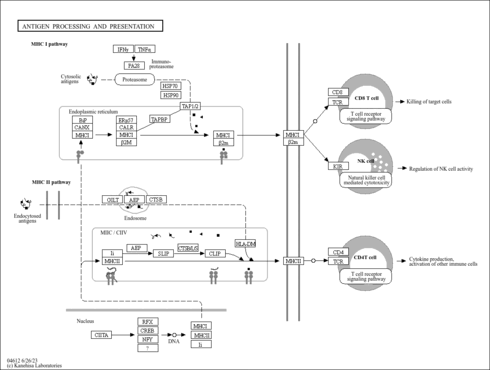
| hsa04612 | Antigen processing and presentation |
|
|---|
| Network |
nt06160 Human T-cell leukemia virus 1 (HTLV-1) nt06161 Human immunodeficiency virus 1 (HIV-1) nt06164 Kaposi sarcoma-associated herpesvirus (KSHV) nt06165 Epstein-Barr virus (EBV) nt06166 Human papillomavirus (HPV) nt06167 Human cytomegalovirus (HCMV) nt06168 Herpes simplex virus 1 (HSV-1) nt06229 MHC presentation (cancer) |
|---|
| Element |
| N00363 | Antigen processing and presentation by MHC class I molecules |
| N00590 | Antigen processing and presentation by MHC class II molecules |
|
|---|
| Drug |
| D03441 | Certolizumab pegol (USAN/INN) |
| D04318 | Glatiramer acetate (JAN/USAN) |
| D12778 | Olgotrelvir sodium (USAN) |
|
|---|
| Other DBs |
|
|---|
| Organism |
Homo sapiens (human) [GN: hsa]
|
|---|
| Gene |
| 100132285 | KIR2DS2; killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 2 [KO:K07982] |
| 10197 | PSME3; proteasome activator subunit 3 [KO:K06698] |
| 10437 | IFI30; IFI30 lysosomal thiol reductase [KO:K08059] |
| 115653 | KIR3DL3; killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 3 [KO:K07980] |
| 1385 | CREB1; cAMP responsive element binding protein 1 [KO:K05870] |
| 3105 | HLA-A; major histocompatibility complex, class I, A [KO:K06751] |
| 3106 | HLA-B; major histocompatibility complex, class I, B [KO:K06751] |
| 3107 | HLA-C; major histocompatibility complex, class I, C [KO:K06751] |
| 3108 | HLA-DMA; major histocompatibility complex, class II, DM alpha [KO:K06752] |
| 3109 | HLA-DMB; major histocompatibility complex, class II, DM beta [KO:K06752] |
| 3111 | HLA-DOA; major histocompatibility complex, class II, DO alpha [KO:K06752] |
| 3112 | HLA-DOB; major histocompatibility complex, class II, DO beta [KO:K06752] |
| 3113 | HLA-DPA1; major histocompatibility complex, class II, DP alpha 1 [KO:K06752] |
| 3115 | HLA-DPB1; major histocompatibility complex, class II, DP beta 1 [KO:K06752] |
| 3117 | HLA-DQA1; major histocompatibility complex, class II, DQ alpha 1 [KO:K06752] |
| 3118 | HLA-DQA2; major histocompatibility complex, class II, DQ alpha 2 [KO:K06752] |
| 3119 | HLA-DQB1; major histocompatibility complex, class II, DQ beta 1 [KO:K06752] |
| 3120 | HLA-DQB2; major histocompatibility complex, class II, DQ beta 2 [KO:K06752] |
| 3122 | HLA-DRA; major histocompatibility complex, class II, DR alpha [KO:K06752] |
| 3123 | HLA-DRB1; major histocompatibility complex, class II, DR beta 1 [KO:K06752] |
| 3125 | HLA-DRB3; major histocompatibility complex, class II, DR beta 3 [KO:K06752] |
| 3126 | HLA-DRB4; major histocompatibility complex, class II, DR beta 4 [KO:K06752] |
| 3127 | HLA-DRB5; major histocompatibility complex, class II, DR beta 5 [KO:K06752] |
| 3133 | HLA-E; major histocompatibility complex, class I, E [KO:K06751] |
| 3134 | HLA-F; major histocompatibility complex, class I, F [KO:K06751] |
| 3135 | HLA-G; major histocompatibility complex, class I, G [KO:K06751] |
| 3303 | HSPA1A; heat shock protein family A (Hsp70) member 1A [KO:K03283] |
| 3304 | HSPA1B; heat shock protein family A (Hsp70) member 1B [KO:K03283] |
| 3305 | HSPA1L; heat shock protein family A (Hsp70) member 1 like [KO:K03283] |
| 3306 | HSPA2; heat shock protein family A (Hsp70) member 2 [KO:K26598] |
| 3308 | HSPA4; heat shock protein family A (Hsp70) member 4 [KO:K09489] |
| 3310 | HSPA6; heat shock protein family A (Hsp70) member 6 [KO:K03283] |
| 3312 | HSPA8; heat shock protein family A (Hsp70) member 8 [KO:K03283] |
| 3320 | HSP90AA1; heat shock protein 90 alpha family class A member 1 [KO:K04079] |
| 3326 | HSP90AB1; heat shock protein 90 alpha family class B member 1 [KO:K04079] |
| 3802 | KIR2DL1; killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 1 [KO:K07981] |
| 3803 | KIR2DL2; killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 2 [KO:K07981] |
| 3804 | KIR2DL3; killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 3 [KO:K07981] |
| 3805 | KIR2DL4; killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 4 [KO:K24231] |
| 3806 | KIR2DS1; killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 1 [KO:K07982] |
| 3808 | KIR2DS3; killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 3 [KO:K07982] |
| 3809 | KIR2DS4; killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 4 (gene/pseudogene) [KO:K07982] |
| 3810 | KIR2DS5; killer cell immunoglobulin like receptor, two Ig domains and short cytoplasmic tail 5 [KO:K07982] |
| 3811 | KIR3DL1; killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 1 [KO:K07980] |
| 3812 | KIR3DL2; killer cell immunoglobulin like receptor, three Ig domains and long cytoplasmic tail 2 [KO:K07980] |
| 3813 | KIR3DS1; killer cell immunoglobulin like receptor, three Ig domains and short cytoplasmic tail 1 [KO:K07980] |
| 3821 | KLRC1; killer cell lectin like receptor C1 [KO:K06541] |
| 3822 | KLRC2; killer cell lectin like receptor C2 [KO:K24233] |
| 3823 | KLRC3; killer cell lectin like receptor C3 [KO:K24233] |
| 3824 | KLRD1; killer cell lectin like receptor D1 [KO:K06516] |
| 4261 | CIITA; class II major histocompatibility complex transactivator [KO:K08060] [EC:2.7.11.1] |
| 4800 | NFYA; nuclear transcription factor Y subunit alpha [KO:K08064] |
| 4801 | NFYB; nuclear transcription factor Y subunit beta [KO:K08065] |
| 4802 | NFYC; nuclear transcription factor Y subunit gamma [KO:K08066] |
| 5720 | PSME1; proteasome activator subunit 1 [KO:K06696] |
| 5721 | PSME2; proteasome activator subunit 2 [KO:K06697] |
| 57292 | KIR2DL5A; killer cell immunoglobulin like receptor, two Ig domains and long cytoplasmic tail 5A [KO:K24232] |
| 5994 | RFXAP; regulatory factor X associated protein [KO:K08063] |
| 6890 | TAP1; transporter 1, ATP binding cassette subfamily B member [KO:K05653] [EC:7.4.2.14] |
| 6891 | TAP2; transporter 2, ATP binding cassette subfamily B member [KO:K05654] [EC:7.4.2.14] |
| 8302 | KLRC4; killer cell lectin like receptor C4 [KO:K24234] |
| 8625 | RFXANK; regulatory factor X associated ankyrin containing protein [KO:K08062] |
|
|---|
| Reference |
|
|---|
| Authors |
Trombetta ES, Mellman I. |
|---|
| Title |
Cell biology of antigen processing in vitro and in vivo. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Inaba K, Inaba M. |
|---|
| Title |
Antigen recognition and presentation by dendritic cells. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Heath WR, Carbone FR. |
|---|
| Title |
Cross-presentation in viral immunity and self-tolerance. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Abele R, Tampe R. |
|---|
| Title |
The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Reith W, LeibundGut-Landmann S, Waldburger JM. |
|---|
| Title |
Regulation of MHC class II gene expression by the class II transactivator. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Honey K, Rudensky AY. |
|---|
| Title |
Lysosomal cysteine proteases regulate antigen presentation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Sercarz EE, Maverakis E. |
|---|
| Title |
Mhc-guided processing: binding of large antigen fragments. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ackerman AL, Cresswell P. |
|---|
| Title |
Cellular mechanisms governing cross-presentation of exogenous antigens. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cresswell P, Ackerman AL, Giodini A, Peaper DR, Wearsch PA. |
|---|
| Title |
Mechanisms of MHC class I-restricted antigen processing and cross-presentation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zika E, Ting JP. |
|---|
| Title |
Epigenetic control of MHC-II: interplay between CIITA and histone-modifying enzymes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Reith W, Mach B. |
|---|
| Title |
The bare lymphocyte syndrome and the regulation of MHC expression. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lizee G, Basha G, Jefferies WA. |
|---|
| Title |
Tails of wonder: endocytic-sorting motifs key for exogenous antigen presentation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Watts C. |
|---|
| Title |
The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kaufmann SH, Schaible UE. |
|---|
| Title |
Antigen presentation and recognition in bacterial infections. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Groettrup M, Kirk CJ, Basler M |
|---|
| Title |
Proteasomes in immune cells: more than peptide producers? |
|---|
| Journal |
|
|---|
Related
pathway |
| hsa04650 | Natural killer cell mediated cytotoxicity |
| hsa04660 | T cell receptor signaling pathway |
|
|---|
| KO pathway |
|
|---|