| Entry |
|
|---|
| Name |
Staurosporine biosynthesis |
|---|
| Class |
Metabolism; Biosynthesis of other secondary metabolites
|
|---|
| Pathway map |
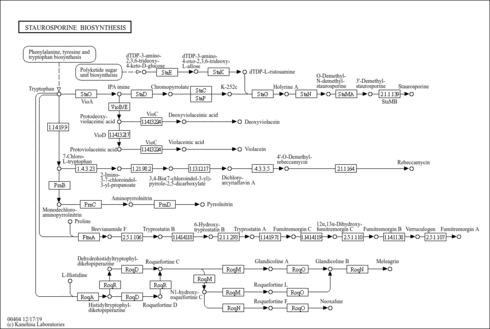
|
|---|
| Module |
| M00786 | Fumitremorgin alkaloid biosynthesis, tryptophan + proline => fumitremorgin C/A [PATH:ec00404] |
| M00789 | Rebeccamycin biosynthesis, tryptophan => rebeccamycin [PATH:ec00404] |
| M00790 | Pyrrolnitrin biosynthesis, tryptophan => pyrrolnitrin [PATH:ec00404] |
| M00805 | Staurosporine biosynthesis, tryptophan => staurosporine [PATH:ec00404] |
| M00808 | Violacein biosynthesis, tryptophan => violacein [PATH:ec00404] |
|
|---|
| Enzyme |
|
|---|
| Compound |
| C07349 | 3'-Demethylstaurosporine |
| C12318 | dTDP-3-amino-2,3,6-trideoxy-4-keto-D-glucose |
| C19688 | 2-Imino-3-(7-chloroindol-3-yl)propanoate |
| C19698 | 3,4-Bis(7-chloroindol-3-yl)pyrrole-2,5-dicarboxylate |
| C19699 | Dichloroarcyriaflavin A |
| C19700 | 4'-O-Demethylrebeccamycin |
| C20513 | 6-Hydroxytryprostatin B |
| C20605 | 12alpha,13alpha-Dihydroxyfumitremorgin C |
| C21110 | Monodechloroaminopyrrolnitrin |
| C21128 | O-Demethyl-N-demethyl-staurosporine |
| C21131 | Protodeoxyviolaceinic acid |
| C21137 | dTDP-3-amino-4-oxo-2,3,6-trideoxy-L-allose |
| C22162 | Histidyltryptophyldiketopiperazine |
| C22164 | Dehydrohistidyltryptophyldiketopiperazine |
| C22169 | N1-Hydroxy-roquefortine C |
|
|---|
| Reference |
|
|---|
| Authors |
Sanchez C, Mendez C, Salas JA |
|---|
| Title |
Indolocarbazole natural products: occurrence, biosynthesis, and biological activity. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Onaka H, Taniguchi S, Igarashi Y, Furumai T |
|---|
| Title |
Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lee ME, Aswani A, Han AS, Tomlin CJ, Dueber JE |
|---|
| Title |
Expression-level optimization of a multi-enzyme pathway in the absence of a high-throughput assay. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Onaka H, Taniguchi S, Igarashi Y, Furumai T |
|---|
| Title |
Characterization of the biosynthetic gene cluster of rebeccamycin from Lechevalieria aerocolonigenes ATCC 39243. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hammer PE, Hill DS, Lam ST, Van Pee KH, Ligon JM |
|---|
| Title |
Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mundt K, Wollinsky B, Ruan HL, Zhu T, Li SM |
|---|
| Title |
Identification of the verruculogen prenyltransferase FtmPT3 by a combination of chemical, bioinformatic and biochemical approaches. |
|---|
| Journal |
|
|---|
Related
pathway |
| ec00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| ec00523 | Polyketide sugar unit biosynthesis |
|
|---|
| LinkDB |
|
|---|