 | | PATHWAY: rn00360 | |
| Entry |
|
|---|
| Name |
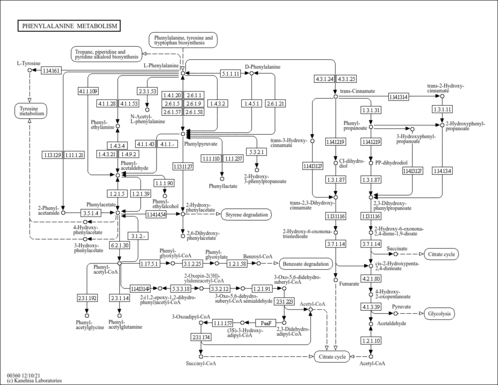
Phenylalanine metabolism |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| M00545 | Trans-cinnamate degradation, trans-cinnamate => acetyl-CoA [PATH:rn00360] |
| M00878 | Phenylacetate degradation, phenylaxetate => acetyl-CoA/succinyl-CoA [PATH:rn00360] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00228 | acetaldehyde:NAD+ oxidoreductase (CoA-acetylating) |
| R00686 | L-phenylalanine racemase (ATP-hydrolysing) |
| R00688 | L-phenylalanine:NAD+ oxidoreductase (deaminating) |
| R00689 | L-phenylalanine:oxygen oxidoreductase (deaminating) |
| R00690 | L-phenylalanine:oxygen 2-oxidoreductase (decarboxylating) |
| R00692 | L-phenylalanine:pyruvate aminotransferase |
| R00693 | acetyl-CoA:L-phenylalanine N-acetyltransferase |
| R00694 | L-phenylalanine:2-oxoglutarate aminotransferase |
| R00697 | L-phenylalanine ammonia-lyase (trans-cinnamate-forming) |
| R00698 | L-phenylalanine:oxygen oxidoreductase (decarboxylating) |
| R00699 | L-phenylalanine carboxy-lyase (phenylethylamine-forming) |
| R00750 | 4-hydroxy-2-oxopentanoate pyruvate-lyase (acetaldehyde-forming) |
| R00829 | succinyl-CoA:acetyl-CoA C-acyltransferase |
| R01370 | phenyllactate:NAD+ oxidoreductase |
| R01371 | phenyllactate:NADP+ oxidoreductase |
| R01372 | phenylpyruvate:oxygen oxidoreductase (hydroxylating,decarboxylating) |
| R01374 | D-phenylalanine:quinone oxidoreductase (deaminating) |
| R01377 | phenylpyruvate carboxy-lyase (phenylacetaldehyde-forming) |
| R01378 | phenylpyruvate keto-enol-isomerase |
| R01582 | D-phenylalanine:2-oxoglutarate aminotransferase |
| R01795 | L-phenylalanine,tetrahydrobiopterin:oxygen oxidoreductase (4-hydroxylating) |
| R02252 | phenylpropanoate:NAD+ delta2-oxidoreductase |
| R02254 | trans-cinnamate,NADPH:oxygen oxidoreductase (2-hydroxylating) |
| R02450 | phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating) |
| R02536 | phenylacetaldehyde:NAD+ oxidoreductase |
| R02537 | aldehyde:NADP+ oxidoreductase |
| R02539 | phenylacetate:CoA ligase |
| R02540 | 2-phenylacetamide amidohydrolase |
| R02576 | phenylacetyl-CoA:L-glutamine alpha-N-phenylacetyltransferase |
| R02601 | 4-hydroxy-2-oxopentanoate hydro-lyase (2-oxopent-4-enoate-forming) |
| R02603 | 2-hydroxy-6-oxonona-2,4-diene-1,9-dioate succinylhydrolase |
| R02611 | phenylethylalcohol:NAD+ oxidoreductase |
| R02612 | phenethylamine:(acceptor) oxidoreductase (deaminating) |
| R02613 | phenethylamine:oxygen oxidoreductase (deaminating) |
| R03369 | 3-(2-hydroxyphenyl)propanoate,NADH2:oxygen oxidoreductase(3-hydroxylating) |
| R03709 | 3-(2-hydroxyphenyl)propanoate:NAD+ oxidoreductase |
| R04376 | 3-(2,3-dihydroxyphenyl)propanoate:oxygen 1,2-oxidoreductase (decyclizing) |
| R05487 | phenylacetate,[reduced NADPH---hemoprotein reductase]:oxygen oxidoreductase (2-hydroxylating) |
| R05841 | phenylacetyl-CoA:glycine N-phenylacetyltransferase |
| R06782 | 3-phenylpropanoate,NADH:oxygen oxidoreductase (2,3-hydroxylating) |
| R06783 | trans-cinnamate,NADH:oxygen oxidoreductase (2,3-hydroxylating) |
| R06784 | 3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)propanoate:NAD+ oxidoreductase |
| R06785 | (2E)-3-(cis-5,6-dihydroxycyclohexa-1,3-dien-1-yl)prop-2-enoate:NAD+ oxidoreductase |
| R06786 | 3-(3-hydroxyphenyl)propanoate,NADH:oxygen oxidoreductase (2-hydroxylating) |
| R06787 | (2E)-3-(3-hydroxyphenyl)prop-2-enoate,NADH:oxygen oxidoreductase (2-hydroxylating) |
| R06788 | (2E)-3-(2,3-dihydroxyphenyl)prop-2-enoate:oxygen 1,2-oxidoreductase (decyclizing) |
| R06789 | (2Z,4E,7E)-2-hydroxy-6-oxonona-2,4,7-triene-1,9-dioate fumarylhydrolase |
| R06941 | (S)-3-hydroxyacyl-CoA:NAD+ oxidoreductase |
| R06942 | (3S)-3-hydroxyacyl-CoA hydro-lyase |
| R07222 | phenylacetyl-CoA:quinone oxidoreductase |
| R07294 | phenylglyoxylyl-CoA hydrolase |
| R09820 | 3-oxo-5,6-dehydrosuberyl-CoA semialdehyde:NADP+ oxidoreductase |
| R09836 | 2-oxepin-2(3H)-ylideneacetyl-CoA hydrolyase |
| R09837 | 2-(1,2-epoxy-1,2-dihydrophenyl)acetyl-CoA isomerase |
| R09838 | phenylacetyl-CoA:oxygen oxidoreductase (1,2-epoxidizing) |
| R09839 | 2,3-didehydroadipoyl-CoA:acetyl-CoA C-didehydroadipoyltransferase (double bond migration) |
| R11918 | L-phenylalanine carboxy-lyase (oxidative-deaminating) |
|
|---|
| Compound |
| C00596 | 2-Hydroxy-2,4-pentadienoate |
| C01198 | 3-(2-Hydroxyphenyl)propanoate |
| C01772 | trans-2-Hydroxycinnamate |
| C02137 | alpha-Oxo-benzeneacetic acid |
| C02763 | 2-Hydroxy-3-phenylpropenoate |
| C03519 | N-Acetyl-L-phenylalanine |
| C03589 | 4-Hydroxy-2-oxopentanoate |
| C04044 | 3-(2,3-Dihydroxyphenyl)propanoate |
| C04479 | 2-Hydroxy-6-oxonona-2,4-diene-1,9-dioate |
| C06207 | 2,6-Dihydroxyphenylacetate |
| C11457 | 3-(3-Hydroxyphenyl)propanoic acid |
| C11588 | cis-3-(Carboxy-ethyl)-3,5-cyclo-hexadiene-1,2-diol |
| C12621 | trans-3-Hydroxycinnamate |
| C12622 | cis-3-(3-Carboxyethenyl)-3,5-cyclohexadiene-1,2-diol |
| C12623 | trans-2,3-Dihydroxycinnamate |
| C12624 | 2-Hydroxy-6-ketononatrienedioate |
| C14144 | 5-Carboxy-2-pentenoyl-CoA |
| C14145 | (3S)-3-Hydroxyadipyl-CoA |
| C19945 | 3-Oxo-5,6-dehydrosuberyl-CoA |
| C19946 | 3-Oxo-5,6-dehydrosuberyl-CoA semialdehyde |
| C19975 | 2-Oxepin-2(3H)-ylideneacetyl-CoA |
| C20062 | 2-(1,2-Epoxy-1,2-dihydrophenyl)acetyl-CoA |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Arai H, Yamamoto T, Ohishi T, Shimizu T, Nakata T, Kudo T. |
|---|
| Title |
Genetic organization and characteristics of the 3-(3-hydroxyphenyl)propionic acid degradation pathway of Comamonas testosteroni TA441. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Diaz E, Ferrandez A, Garcia JL. |
|---|
| Title |
Characterization of the hca cluster encoding the dioxygenolytic pathway for initial catabolism of 3-phenylpropionic acid in Escherichia coli K-12. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ferrandez A, Garcia JL, Diaz E. |
|---|
| Title |
Genetic characterization and expression in heterologous hosts of the 3-(3-hydroxyphenyl)propionate catabolic pathway of Escherichia coli K-12. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Hwang JY, Park J, Seo JH, Cha M, Cho BK, Kim J, Kim BG |
|---|
| Title |
Simultaneous synthesis of 2-phenylethanol and L-homophenylalanine using aromatic transaminase with yeast Ehrlich pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Teufel R, Mascaraque V, Ismail W, Voss M, Perera J, Eisenreich W, Haehnel W, Fuchs G |
|---|
| Title |
Bacterial phenylalanine and phenylacetate catabolic pathway revealed. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| rn00960 | Tropane, piperidine and pyridine alkaloid biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|