 | | PATHWAY: rn00470 | |
| Entry |
|
|---|
| Name |
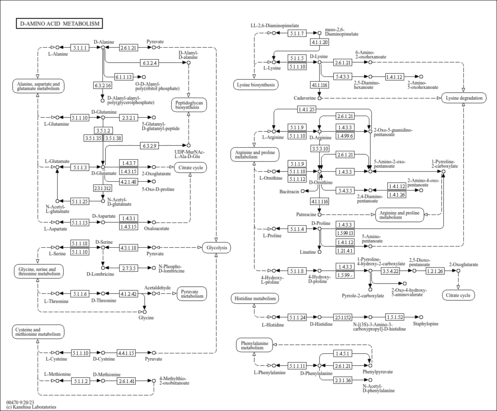
D-Amino acid metabolism |
|---|
| Class |
Metabolism; Metabolism of other amino acids
|
|---|
| Pathway map |
|
|---|
| Module |
| M00947 | D-Arginine racemization, D-arginine => L-arginine [PATH:rn00470] |
| M00948 | Hydroxyproline degradation, trans-4-hydroxy-L-proline => 2-oxoglutarate [PATH:rn00470] |
| M00949 | Staphylopine biosynthesis, L-histidine => staphylopine [PATH:rn00470] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00264 | 2,5-dioxopentanoate:NADP+ 5-oxidoreductase |
| R00279 | D-glutamate:oxygen oxidoreductase (deaminating) |
| R00359 | D-aspartate:oxygen oxidoreductase (deaminating) |
| R00451 | meso-2,6-diaminoheptanedioate carboxy-lyase (L-lysine-forming) |
| R00686 | L-phenylalanine racemase (ATP-hydrolysing) |
| R01148 | D-alanine:2-oxoglutarate aminotransferase |
| R01150 | D-alanine:D-alanine ligase (ADP-forming) |
| R01344 | D-alanine:2-oxo acid aminotransferase |
| R01374 | D-phenylalanine:quinone oxidoreductase (deaminating) |
| R01579 | D-glutamine amidohydrolase |
| R01582 | D-phenylalanine:2-oxoglutarate aminotransferase |
| R01583 | D-glutamate hydro-lyase (cyclizing; 5-oxo-D-proline-forming) |
| R01874 | D-cysteine sulfide-lyase (deaminating; pyruvate-forming) |
| R02280 | 1-pyrroline-4-hydroxy-2-carboxylate aminohydrolase (decyclizing) |
| R02457 | D-ornithine:oxygen oxidoreductase (deaminating) |
| R02458 | D-arginine amidinohydrolase |
| R02459 | D-ornithine:2-oxoglutarate aminotransferase |
| R02460 | bacitracin synthetase 2 |
| R02461 | D-ornithine 4,5-aminomutase |
| R02718 | D-alanine:poly(ribitol phosphate) ligase (AMP-forming) |
| R02735 | LL-2,6-diaminoheptanedioate 2-epimerase |
| R02783 | UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase (ADP-forming) |
| R02851 | D-lysine:2-oxocarboxylate aminotransferase |
| R02852 | D-2,6-diaminohexanoic acid 5,6-aminomutase |
| R02894 | D-proline:oxygen oxidoreductase |
| R02923 | D-arginine:oxygen oxidoreductase (deaminating) |
| R02924 | D-arginine:2-oxoglutarate aminotransferase |
| R03001 | D-methionine:pyruvate aminotransferase |
| R03296 | trans-4-hydroxy-L-proline 2-epimerase |
| R03691 | ATP:lombricine N-phosphotransferase |
| R03903 | acetyl-CoA:D-amino-acid N-acetyltransferase |
| R04012 | D-glutamine:D-glutamyl-peptide 5-glutamyltransferase |
| R04200 | D-threo-2,4-diaminopentanoate:NAD+ oxidoreductase (deaminating) |
| R04201 | D-threo-2,4-diaminopentanoate:NADP+ oxidoreductase(deaminating) |
| R04221 | cis-4-hydroxy-D-proline:oxygen oxidoreductase (deaminating) |
| R04369 | D-alanine:alanyl-poly(glycerolphosphate) ligase (ADP-forming) |
| R04687 | 2,5-diaminohexanoate:NAD+ oxidoreductase (deaminating) |
| R04688 | 2,5-diaminohexanoate:NADP+ oxidoreductase (deaminating) |
| R08195 | D-threonine acetaldehyde-lyase (glycine-forming) |
| R09496 | D-proline:acceptor oxidoreductase |
| R11018 | D-arginine:acceptor oxidoreductase (deaminating) |
| R11031 | L-arginine:NAD+ oxidoreductase (deaminating) |
| R11032 | L-arginine:NADP+ oxidoreductase (deaminating) |
| R11428 | cis-4-hydroxy-D-proline:acceptor oxidoreductase |
| R12237 | staphylopine:NADP+ oxidoreductase [(2S)-2-amino-4-{[(1R)-1-carboxy-2-(1H-imidazol-4-yl)ethyl]amino}butanoate]-forming |
| R12304 | D-ornithine carboxy-lyase |
| R12341 | S-adenosyl-L-methionine:D-histidine N-[(3S)-3-amino-3-carboxypropyl]-transferase |
| R13097 | N-acetyl-glutamate racemase |
|
|---|
| Compound |
| C00666 | LL-2,6-Diaminoheptanedioate |
| C00680 | meso-2,6-Diaminoheptanedioate |
| C00692 | UDP-N-acetylmuramoyl-L-alanyl-D-glutamate |
| C01110 | 5-Amino-2-oxopentanoic acid |
| C01180 | 4-Methylthio-2-oxobutanoic acid |
| C03341 | 2-Amino-4-oxopentanoic acid |
| C03440 | cis-4-Hydroxy-D-proline |
| C03564 | 1-Pyrroline-2-carboxylate |
| C03771 | 5-Guanidino-2-oxopentanoate |
| C03933 | 5-D-Glutamyl-D-glutamyl-peptide |
| C03943 | (2R,4S)-2,4-Diaminopentanoate |
| C04260 | O-D-Alanyl-poly(ribitol phosphate) |
| C04282 | 1-Pyrroline-4-hydroxy-2-carboxylate |
| C04457 | D-Alanyl-alanyl-poly(glycerolphosphate) |
| C05161 | (2R,5S)-2,5-Diaminohexanoate |
| C05620 | N-Acetyl-D-phenylalanine |
| C05941 | 2-Oxo-4-hydroxy-5-aminovalerate |
| C22025 | (2S)-2-Amino-4-{[(1R)-1-carboxy-2-(1H-imidazol-4-yl)ethyl]amino}butanoate |
|
|---|
| Reference |
|
|---|
| Authors |
Miyamoto T, Homma H |
|---|
| Title |
D-Amino acid metabolism in bacteria. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ghssein G, Brutesco C, Ouerdane L, Fojcik C, Izaute A, Wang S, Hajjar C, Lobinski R, Lemaire D, Richaud P, Voulhoux R, Espaillat A, Cava F, Pignol D, Borezee-Durant E, Arnoux P |
|---|
| Title |
Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00250 | Alanine, aspartate and glutamate metabolism |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00270 | Cysteine and methionine metabolism |
| rn00330 | Arginine and proline metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|