Reaction Module
KEGG pathway modules in the metabolic pathways represent functional units defined by a set of KO identifiers (K numbers) for the enzymes involved. In contrast, KEGG reaction modules are defined from purely chemical data without incorporating any enzyme data, i.e., from the analysis of chemical structure transformation patterns along the metabolic pathways. The reaction module is a conserved sequence of chemical structure transformation patterns defined by a set of Reaction Class identifiers (RC numbers).
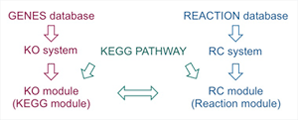
The Reaction Class is like a reaction ortholog accommodating global structural differences of metabolites. Therefore, it is interesting to examine how well reaction modules (RC modules) defined by reaction orthologs correspond to KEGG pathway modules (KO modules) defined by gene orthologs. This is summarized in the following BRITE hierarchy file.
Reaction modules are now part of the MODULE flat file (see KEGG MODULE Database).
Carboxylic acid metabolism
2-Oxocarboxylic acid chain extension
- RM001
- 2-Oxocarboxylic acid chain extension by tricarboxylic acid pathway
2-Oxocarboxylic acid chain modification
- RM002
- Carboxyl to amino conversion using protective N-acetyl group (basic amino acid synthesis)
- RM032
- Carboxyl to amino conversion without using protective group
- RM033
- Branched-chain addition (branched-chain amino acid synthesis)
- RM030
- Glucosinolate synthesis
- RM044
- 2-Oxocarboxylic acid conversion to its CoA derivative
- RM901
- Reductive amination of 2-oxocarboxylic acid (aminotransferase reaction)
- RM903
- Oxidative decarboxylation of 2-oxoacid to form its CoA derivative
Fatty acid synthsis and degradation
Acyl-CoA metabolism
- RM019
- Acyl-CoA conversion via dicarboxylate semialdehyde
Reaction module variation
- RM032 RM002
- Carboxyl to amino conversion
- RM020 RM021
- Fatty acid synthesis
- RM018 RM016
- Beta oxidation
Reaction module combination
- RM001+(RM002,RM032,RM033)
- Amino acid synthesis
- RM001+RM030
- Glucosinolate synthesis
Reaction module map
Nucleotide sugar metabolism
Nucleotide sugar biosynthesis
Deoxy sugar metabolism
- RM035
- Transamination and N,N-dimethylation in polyketide biosynthesis
- RM043
- Transamination and N-acetylation in deoxysugar biosynthesis
- RM036
- Epimerization and reduction in deoxysugar biosynthesis
- RM037
- Deoxygenation (transamination and deamination) in deoxysugar biosynthesis
- RM038
- Deoxygenation (dehydration and reduction) in deoxysugar biosynthesis
Sugar conversion
Reaction module variation
- RM022 RM023 RM039
- Nucleotide sugar biosynthesis (sugar activation)
Reaction module combination
- (RM022,RM039)+(RM040,RM041)
- Ascorbate synthesis
Reaction module map
- map01250
- Biosynthesis of nucleotide sugars
Aromatics degradation
Methyl to carboxyl conversion on aromatic ring (preprocessing module)
Dihydroxylation of aromatic ring
- RM004
- Dihydroxylation of aromatic ring, type 1 (dioxygenase and dehydrogenase reactions)
- RM005
- Dihydroxylation of aromatic ring, type 1a (dioxygenase and decarboxylating dehydrogenase reactions)
- RM902
- Dihydroxylation of aromatic ring, type 1b (single dioxygenase reaction)
- RM006
- Dihydroxylation of aromatic ring, type 2 (two monooxygenase reactions)
- RM007
- Dihydroxylation of aromatic ring, type 3 (dealkylation and monooxygenase reactions)
Cleavage of aromatic ring
Dihydroxylation and cleavage of aromatic ring
Reaction module comparison
- RM003 RM015
- Methyl to carboxyl conversion of aromatic ring
Reaction module combination
- RM003+((RM004,RM005,RM006,RM007)+(RM008,RM009),RM010)
- Aromatics degradation
Reaction module map
- map01220
- Degradation of aromatic compounds
Other
Amino acid metabolism
- RM025
- Conversion of amino acid moiety to carboxyl group (biogenic amine metabolism)
Nucleotide metabolism
Sugar metabolism
- RM034
- Sugar degradation to aldehyde and pyruvate
Aromatics metabolism
References
- Muto, A., Kotera, M., Tokimatsu, T., Nakagawa, Z., Goto, S., and Kanehisa, M.; Modular architecture of metabolic pathways revealed by conserved sequences of reactions. J. Chem. Inf. Model. 53, 613-622 (2013). [pubmed] [pdf]
- Kanehisa, M.; Chemical and genomic evolution of enzyme-catalyzed reaction networks. FEBS Lett. 587, 2731-2737 (2013). [pubmed] [pdf]
- Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M.; Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014). [pubmed] [pdf]
Last updated: September 20, 2023
